Shaping GWAS: Integrating DNA Topology Into Genetic Association Studies
AUTHOR: Ekaterina Khvatkova
PUBLISHED: August 26th, 2025
MODIFIED: October 4th, 2025
Introduction
Genome-wide association studies (GWAS) integrate statistical and biological insights to uncover the genetic basis of complex diseases, relying on functional annotations to facilitate casual variant identification. Despite the availability of high-throughput, laboratory-based functional datasets, many biologically relevant phenomena remain excluded from GWAS pipelines due to technical limitations in database granularity, inherent biases, and the absence of robust methodological frameworks for integration. One key biological phenomenon is the effect of genetic variants on DNA topology.
What is DNA Topology?
Tertiary DNA topology refers to the intrinsic 3D structural features of DNA determined by nucleotide sequences. Although DNA is commonly depicted as a uniform double helix, its three-dimensional helix structure depends on the specific alleles present at a given genomic position. While all genetic variants influence DNA conformation to some extent, certain variants induce more pronounced alterations to its shape.
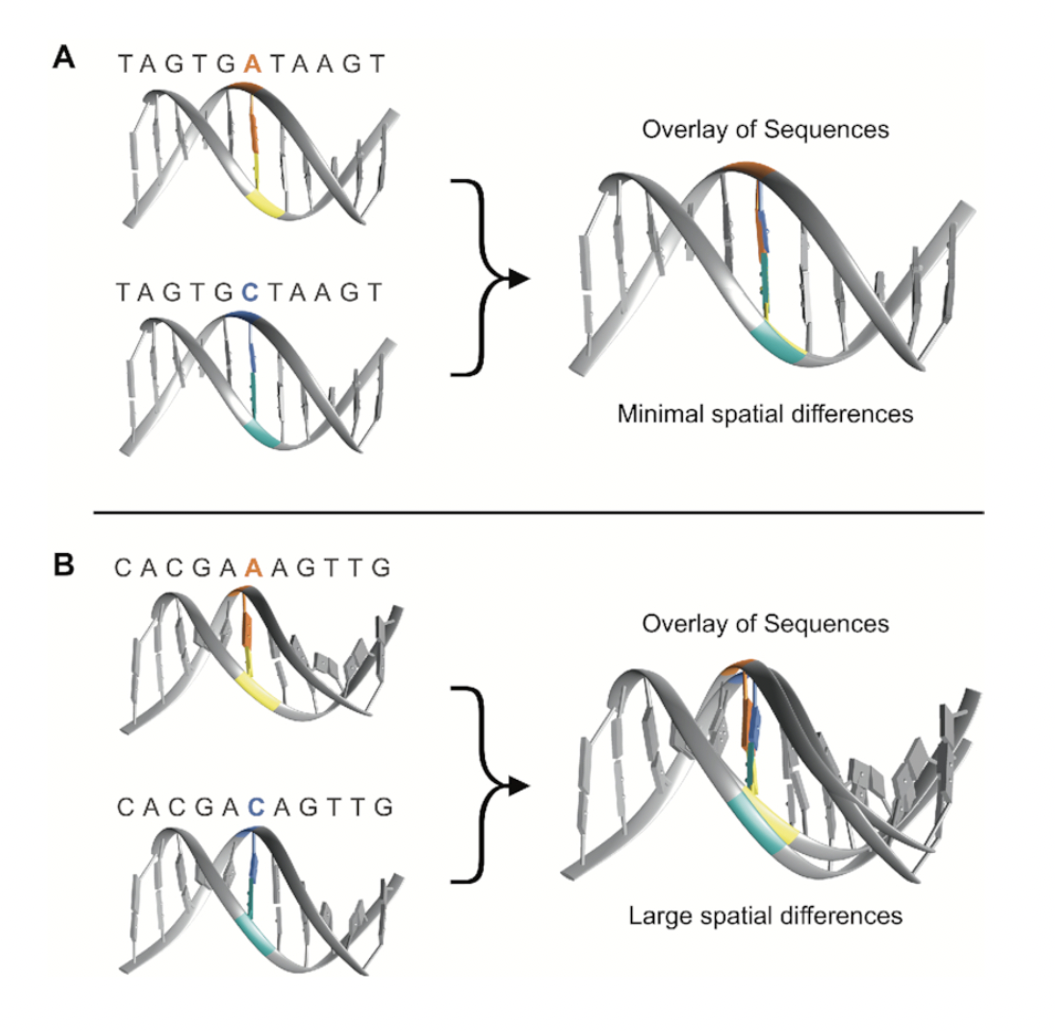
The effect of local sequence context on DNA conformation is illustrated in Figure 1. (A) and (B) both depict sequences containing an A/C single nucleotide variant. However, in contrast to the static DNA conformation observed in (A), (B) reveals substantial alterations in spatial geometry. This discrepancy arises from the influence of the sequence surrounding the genetic variant on DNA shape.
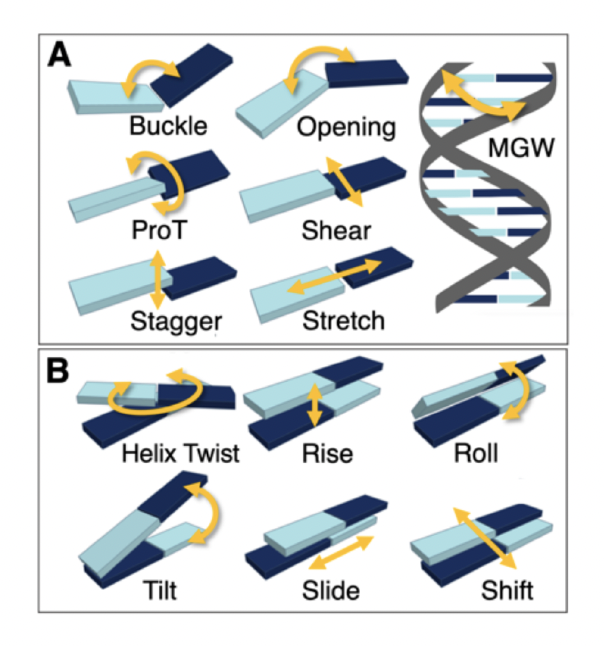
These conformational differences of DNA can be characterized through shape features (illustrated in Figure 2). These shape features describe the local three-dimensional geometry of the DNA molecule at different points along its double helix beyond the standard double helix structure.
In the context of genetic association studies, where statistical models often evaluate the effect of a single genetic variant on a disease-related trait and do not incorporate the sequence surrounding that variant, such structural nuances are traditionally overlooked.
Research Aims
With principal investigators Hannah Ainsworth and Carl Langefeld at Wake Forest University School of Medicine, we are developing and applying a paradigm integrating DNA topology within statistical genomics to refine the identification of causal variants of complex diseases. This foundational work will enable new statistical methods for unraveling the etiology of complex diseases and identifying plausible therapeutic targets.
Acknowledgements
Funding for this work is provided by NIH grants R01 EY030521, 7 R01 AR078785-03, and U01 NS069763, USAMRDC project W81XWH-20-1-0686, and a 2023 Daryl and Marguerite Errett Discovery Award (sponsored by the Errett Fisher Foundation).